install.packages("REDCapTidieR")REDCapTidieR 0.4.0
We’re thrilled to announce the release of REDCapTidieR v0.4.0 on CRAN! REDCapTidieR provides a user-friendly way to import data from a REDCap project into the R environment.
This blog post will introduce you to three brand new features we’re excited to share.
You can install the current version from CRAN with:
Get Metadata Summary Statistics 🔎
The skimr package is a powerful tool allowing users to quickly get summary statistics of their data. REDCapTidieR now provides a simple function that will automatically apply the appropriate metrics to the supertibble metadata using add_skimr_metadata().
Let’s take a look at what this does using the internal superheroes_supertbl dataset by first re-acquainting ourselves with a typical supertibble:
library(REDCapTidieR)
library(dplyr)
superheroes_supertbl |>
rmarkdown::paged_table()Now, let’s apply our new function and observe some pre-selected metadata variables of interest in the redcap_metadata metadata tibbles:
superheroes_supertbl |>
add_skimr_metadata() |>
select(redcap_metadata) |>
tidyr::unnest() |>
select(1, 2, 18:33) |>
rmarkdown::paged_table()In addition to the typical metadata columns, we now see a slew of new columns providing metadata statistics! add_skimr_metadata() will automatically detect and apply the appropriate metrics based on the REDCap metadata.
Curious about what metrics are available? You can find out more using skimr::get_default_skimmer_names() and check out the documentation.
Export Supertibbles to XLSX 📝
One of the most common mediums for data analysis and collaboration is the spreadsheet. With this release, you can now export your supertibble to Excel XLSX using the new write_redcap_xlsx() function. Let’s observe a couple of the output sheets:
superheroes_supertbl |>
write_redcap_xlsx("superheroes.xlsx")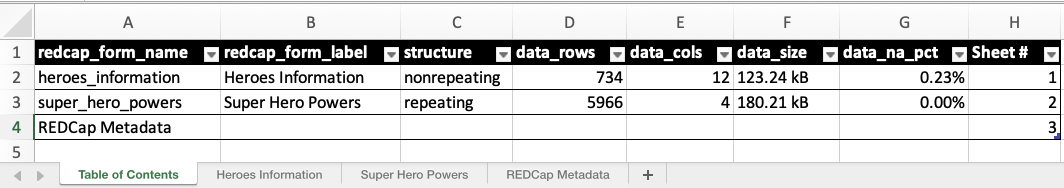
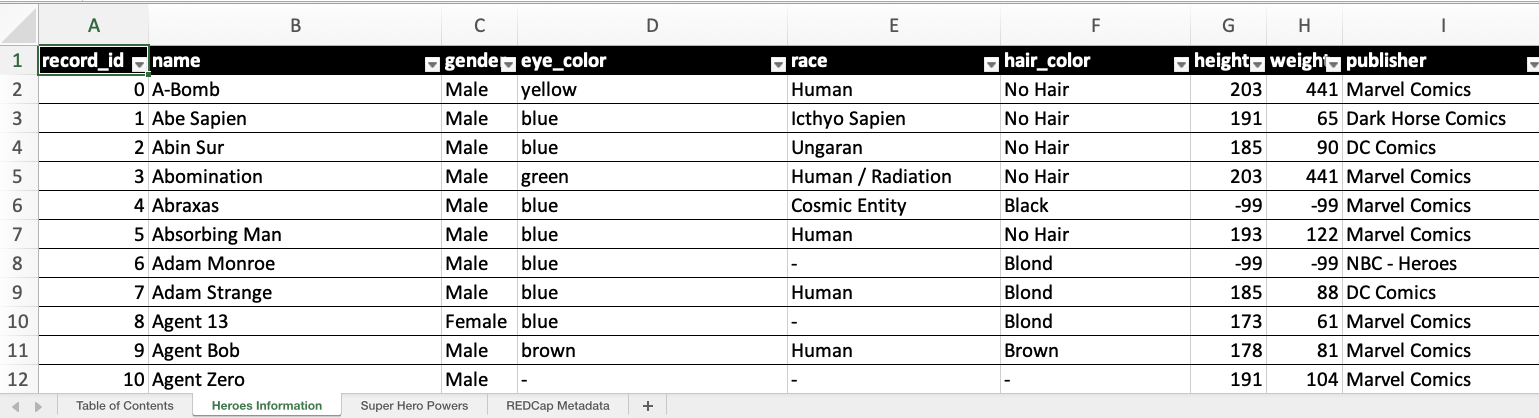
By default, when opening the Excel document you’ll see multiple sheets. The first is the Table of Contents, followed by individual sheets for each instrument’s data, and ending with REDCap Metadata. The Table of Contents provides an overview of data present in the rest of the document while REDCap Metadata provides metadata for all fields from all instruments in one location. Each of these are optional and can be turned off if you only want the data sheets.
Labelled XLSX
As introduced in REDCapTidieR v0.2.0, the labelled package can still be used in your XLSX output files. By using the make_labelled() function, you can now integrate your labels into your Excel sheets:
superheroes %>%
make_labelled() %>%
write_redcap_xlsx("superheroes-labelled.xlsx")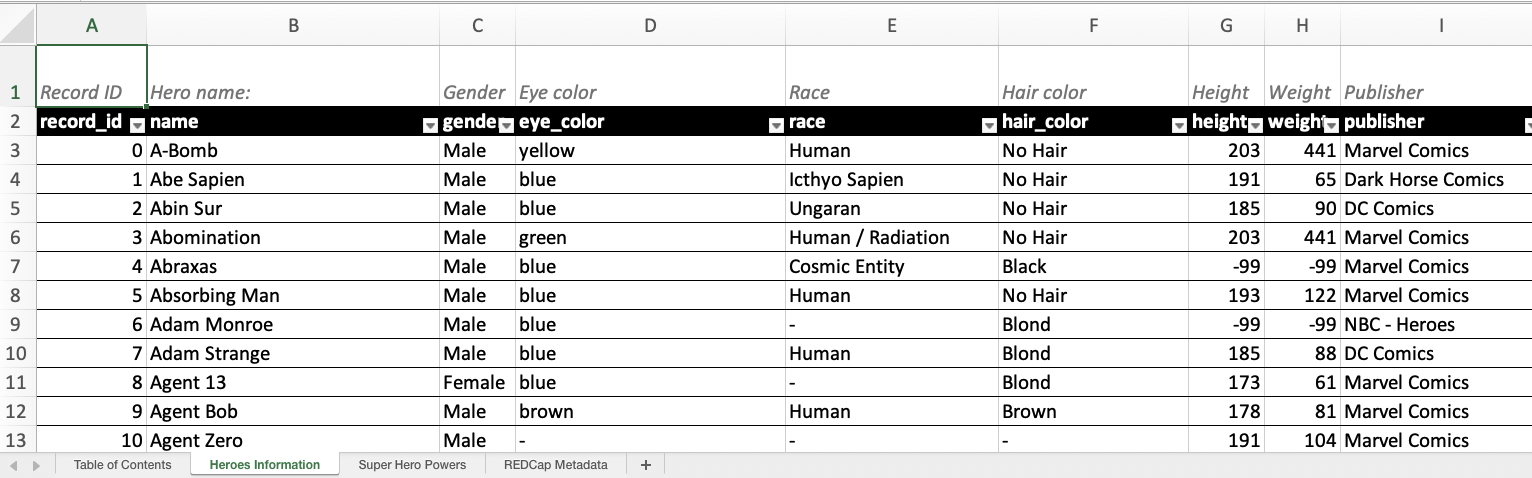 Observe how the first row now contains human-readable labels!
Observe how the first row now contains human-readable labels!
Recoding Logical Columns
By default, write_redcap_xlsx() will recode logical (TRUE/FALSE) columns into more human-friendly terms. This can be adjusted using the recode_logical argument:
superheroes %>%
write_redcap_xlsx("superheroes-labelled.xlsx", recode_logical = TRUE)write_redcap_xlsx() uses the field label to decide how to recode logical fields:
- Columns derived from yesno fields are recoded from
TRUE/FALSEtoyes/no - Columns derived from checkbox fields are recoded from
TRUE/FALSEtoChecked/Unchecked - Columns derived from truefalse fields are left as is (
TRUE/FALSE)
Setting recode_logical to FALSE will preserve all logical fields as (TRUE/FALSE).
Data Access Group Support 🏢
In REDCap projects, Data Access Groups (DAGs) are used to assign user privileges and control record-level access. DAGs are particularly useful in multi-center studies where data for one participant needs to be kept hidden from other participants in the project.
When using the read_redcap() function, DAGs are automatically exported if detected. You can toggle this option with the export_data_access_groups argument. If declared and/or detected, the redcap_data data tibble will be augmented with an additional data column called redcap_data_access_group, which indicates the assigned DAG for each record.
Let’s look at a practical example below:
my_redcap_data <- read_redcap(redcap_uri, dag_token, export_data_access_groups = TRUE)
my_redcap_data |>
extract_tibbles() |>
purrr::pluck(1)# A tibble: 4 × 4
record_id redcap_data_access_group non_repeat_form_1_txt form_status_complete
<dbl> <chr> <lgl> <fct>
1 1 dag1 NA Complete
2 2 dag2 NA Complete
3 3 dag3 NA Complete
4 4 NA NA Incomplete Here, you can see the redcap_data_access_group column identifies which DAG the record belongs to as well as one example where a record wasn’t assigned to any at the bottom.
Bug Fixes
- Fixed a bug where REDCapR API error messages weren’t being returned from REDCapTidieR
Miscellaneous
- All deprecated functions have been officially retired and removed from the package