Excel spreadsheets are an accessible medium for data analysis and collaboration. This vignette will guide you through the process of using REDCapTidieR to transform your REDCap data into an XLSX document, facilitating easy sharing with colleagues.
write_redcap_xlsx() also works with
add_skimr_metadata() to supply both labelled and unlabelled
skimr metadata metrics to your XLSX document.
Transforming a REDCap project into an XLSX document
The transformation of a REDCap project into a XLSX document can be
achieved in three straightforward steps using the
write_redcap_xlsx() function:
- Specify your REDCap URI and API token.
- Invoke
read_redcap(). - Apply
write_redcap_xlsx().
Below is a practical illustration:
redcap_uri <- "https://my.institution.edu/redcap/api/"
token <- "123456789ABCDEF123456789ABCDEF04"
my_redcap_data <- read_redcap(redcap_uri, token)
write_redcap_xlsx(my_redcap_data, file = "my_redcap_data.xlsx")The resulting file will contain the data corresponding to each REDCap instrument in a dedicated sheet. Additionally, a “Table of Contents” sheet will be at the beginning, and a “REDCap Metadata” sheet will be at the end.
Diving Deeper
We will revisit the Superheroes dataset from the Getting Started vignette.
First, we’ll use read_redcap() to create the
superheroes supertibble:
superheroes_token <- "123456789ABCDEF123456789ABCDEF04"
redcap_uri <- "https://my.institution.edu/redcap/api/"
superheroes <- read_redcap(redcap_uri, superheroes_token)
superheroes |>
rmarkdown::paged_table()The superheroes supertibble contains two rows, one for
each instrument of the
Superheroes REDCap project. The
write_redcap_xlsx() function, leveraging the openxlsx2 package,
understands the supertibble’s structure and creates a
collaborator-friendly XLSX file.
Let’s try it out!
superheroes |>
write_redcap_xlsx(file = "superheroes.xlsx")When you open the Excel document, you’ll see multiple sheets. The first one is the Table of Contents sheet, followed by individual sheets for each instrument’s data, and concluded with a REDCap Metadata sheet.
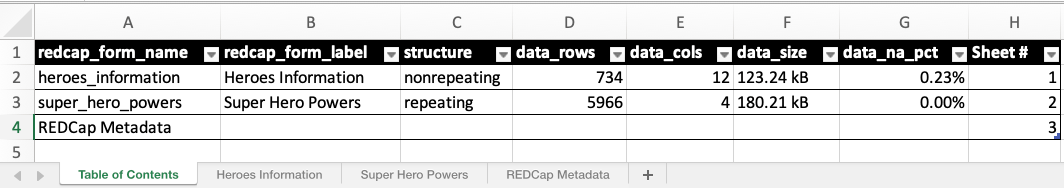
superheroes.xlsx
The Table of Contents sheet provides an overview of
the data present in the rest of the document. If you wish to exclude
this sheet, set the include_toc_sheet argument of
write_redcap_xlsx() to FALSE.
Each instrument’s data is contained in a separate sheet. For a better understanding of how REDCapTidieR represents REDCap data, refer to the Diving Deeper vignette.
The column headers come with pre-applied filters (notice the arrow on the right side of each header cell), making data exploration straightforward.
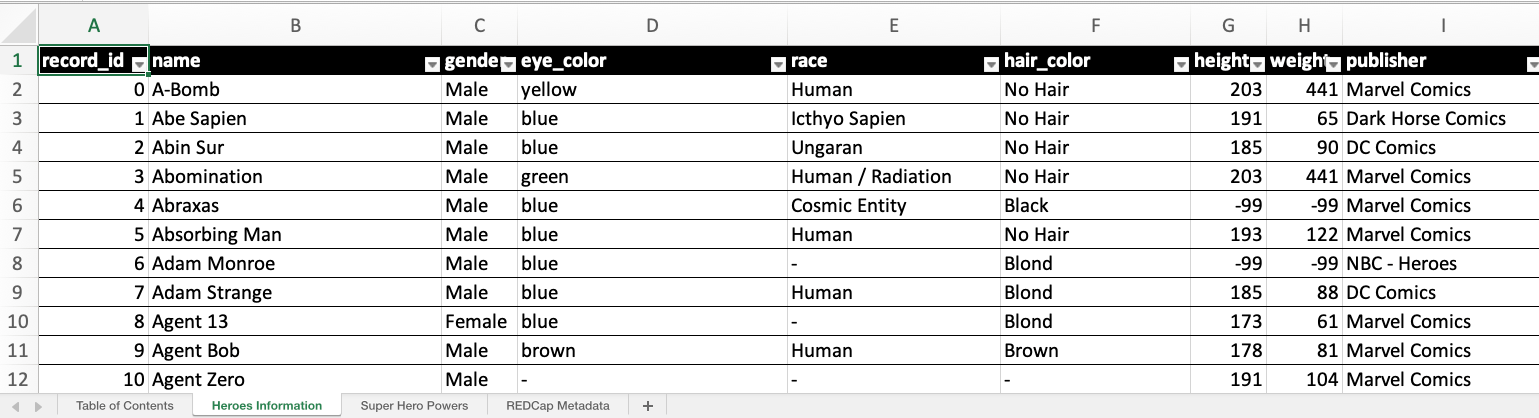
superheroes.xlsx
The REDCap Metadata sheet offers metadata for
all fields from all instruments in one place. If you
don’t want this sheet, set the include_metadata_sheet
argument of write_redcap_xlsx() to FALSE.
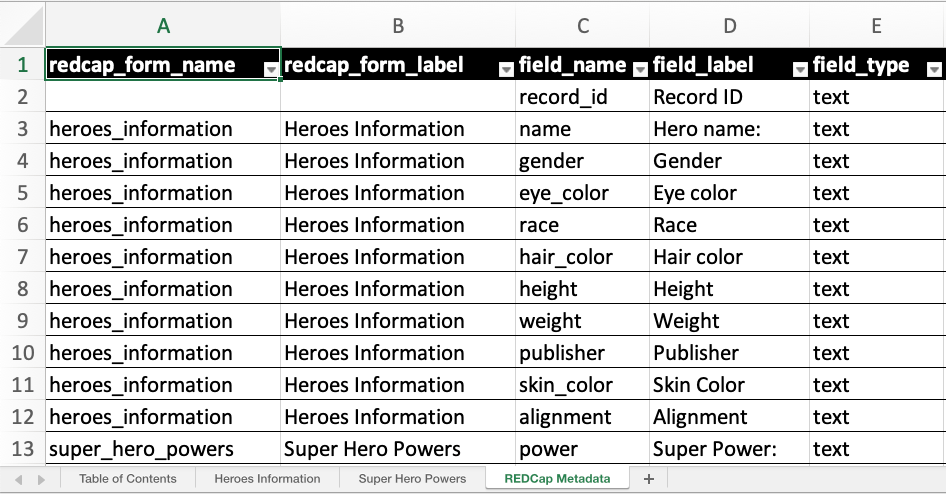
superheroes.xlsx
Applying Labels
Labels can offer valuable context to collaborators, helping them
understand the data in a specific column. The
make_labelled() function allows you to add variable labels to a
REDCapTidieR supertibble, and
write_redcap_xlsx() can integrate these labels to the Excel
sheets.
Let’s add variable labels to superheroes and export it
to Excel:
superheroes |>
make_labelled() |>
write_redcap_xlsx("superheroes-labelled.xlsx")Observe how the first row now contains a human-readable description for each column.
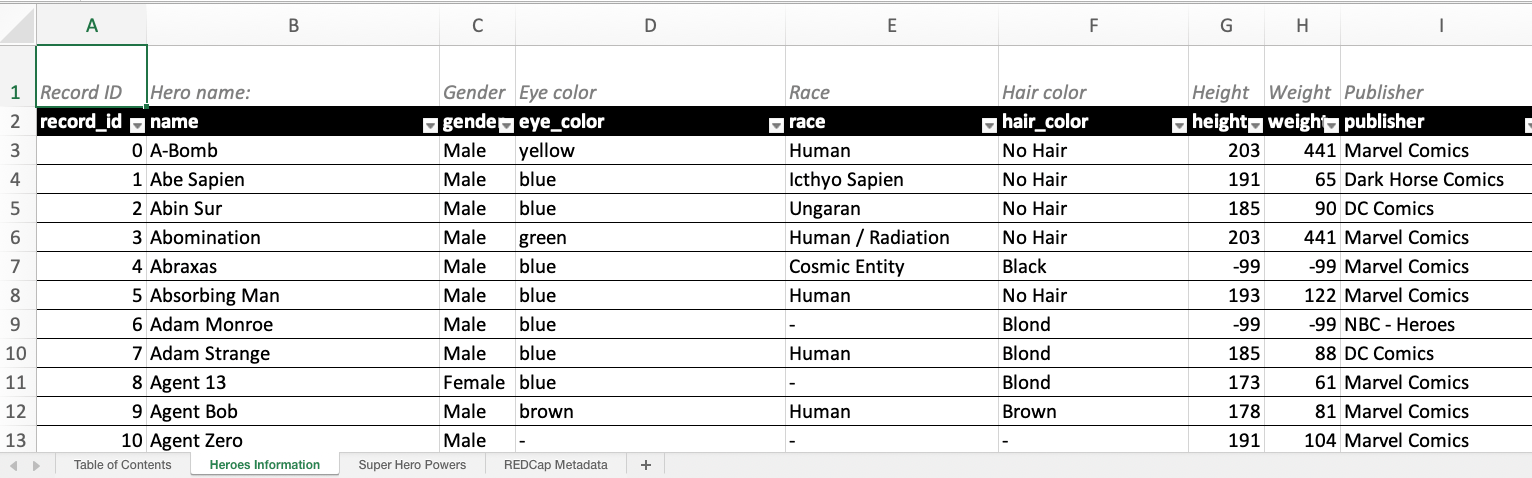
Recoding Logical Columns
By default, write_redcap_xlsx() recodes logical
(TRUE/FALSE) columns into more human-friendly
terms. You can adjust this behavior using the
recode_logical argument:
supertbl |>
write_redcap_xlsx(recode_logical = TRUE)write_redcap_xlsx() uses the field label to decide how to recode
logical fields:
- Columns derived from yesno fields are recoded from
TRUE/FALSEtoyes/no - Columns derived from checkbox fields are recoded
from
TRUE/FALSEtoChecked/Unchecked - Columns derived from truefalse fields are left as
is (
TRUE/FALSE)
Setting recode_logical to FALSE will
preserve all logical fields as
(TRUE/FALSE).
